Walkthrough with real data set ( GSE67959 )
The CapSequm web tool is located in http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi
Via the web tool, you can read the documentation like so :
To make a design, you need to mark your regions of interest to a bed file like so :
chr11 32183395 32183396 Hba-1 chr11 32196221 32196222 Hba-2 chr11 69393434 69393435 TP53 chr11 77886673 77886674 Mir144145
Note how the regions just "mark" the location - 1b wide region is best.
The fourth column is the capture site name. The only allowed characters are alphanumerics (AZaz09), and _- (hyphen, underscore). No whitespace.
Here a ready-made 1b wide region file, for the 30g (30 genes) data set used in the walk-through examples : 30g_forCapSequm.bed
CapSequm will then locate, within which RE fragment this region resides, and will try to design a capture oligo to both ends of this fragment.
You can use UCSC browser to help you locate, where you want to put your capture sites. UCSC browser has a lot of genomics data (RNA expression, histone markers, transcription factors etc) available, to aid you to pinpoint your regions of interest. You can also load in your own data, and flexibly combine that with the data provided by the UCSC browser. The UCSC server addresses are : https://genome-euro.ucsc.edu (European) and https://genome.ucsc.edu (American)
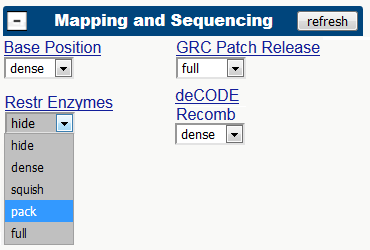
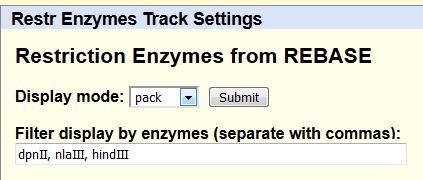
CapSequm will try to place capture oligo to as close to the RE cut site as possible. You can have a look at the RE fragments already when planning for CapSequm run, by turning the "RE enzyme cut sites" track on in UCSC browser :
To plan ahead, you may wish to turn on "repeat masker" track in UCSC browser :
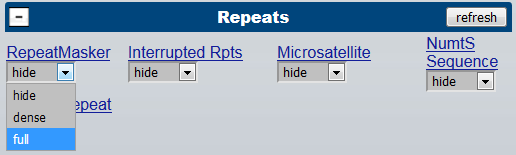
This allows you to avoid repeat rich regions (CapSequm will reject repeat-rich oligo designs, and for these capture sites a new location needs to be chosen, and CapSequm re-ran, to complete the design)
Here a ready-made 1b wide region file, for the 30g (30 genes) data set used in the walk-through examples : 30g_forCapSequm.bed
You can now load the above to the CapSequm web tool, and generate the design : http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi
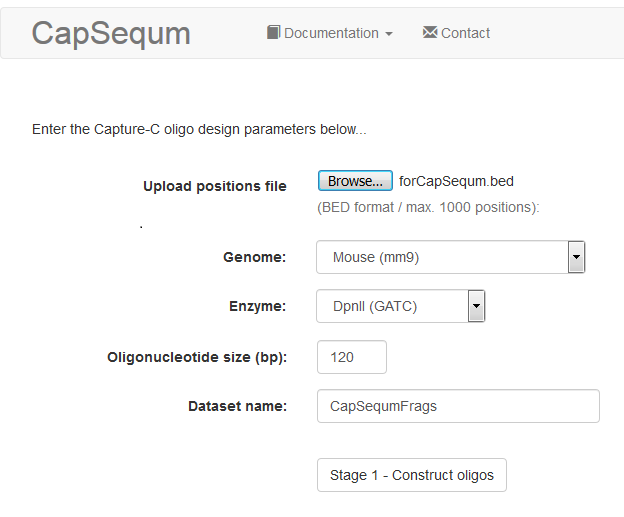
As we are designing for mouse, we need to check that the correct genome is selected when running the CapSequm tool :
In the above :
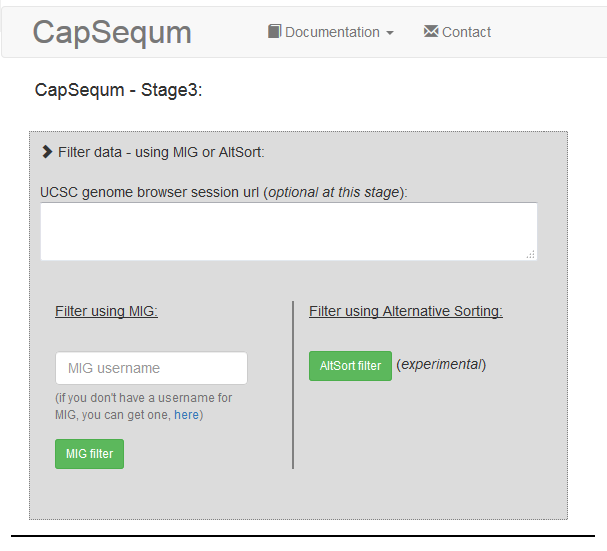
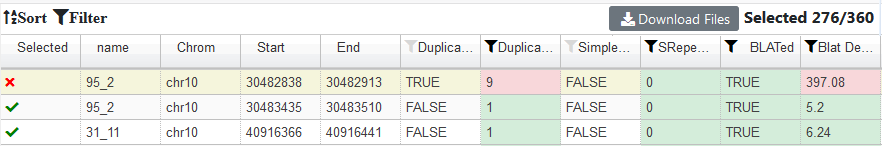
In the end it allows you to filter the data. Select the "Alternative Sorting" - this is easier for external users :
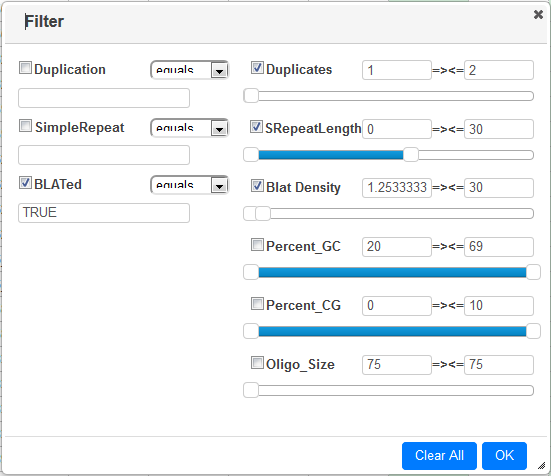
The default filtering parameters are normally fine, but you can finetune them by clicking the Filter button :
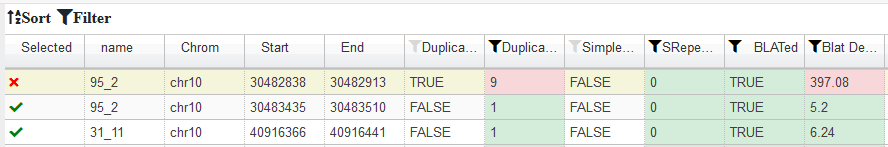
In the end you want to Download Files , to save your design :
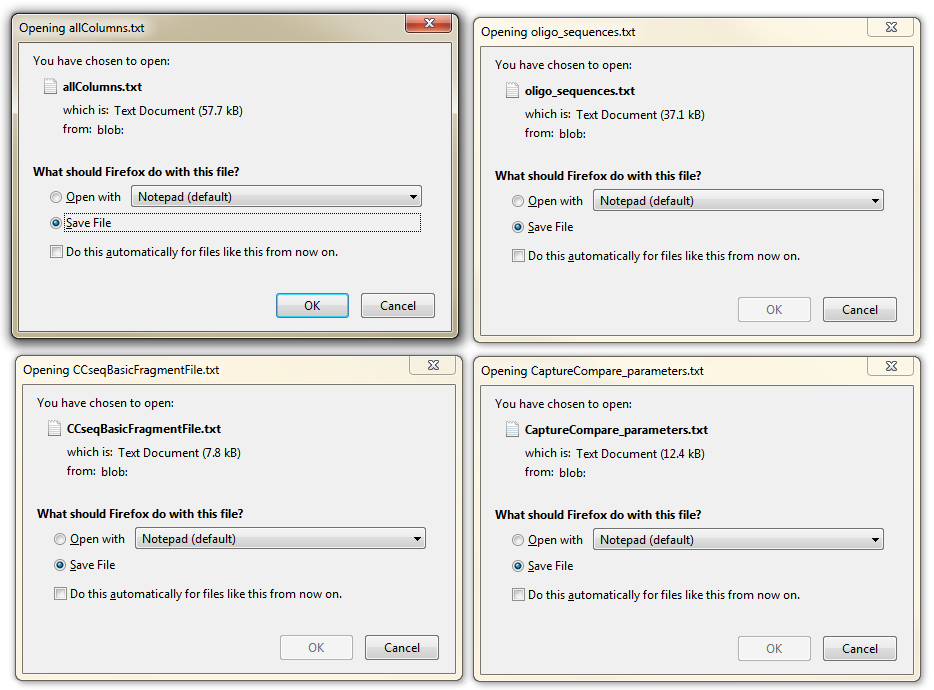
allColumns.txt contains all the designed oligos (also the ones which failed filtering) The passed and failed oligos are marked clearly in this file - serving as a note of the RE fragments that need to be re-designed, as both (or one) RE fragment ends failed in CapSequm.
( For the failed RE fragments, repeat the above protocol, to reach full set of desired capture sites. )
The rest of the files contain only the regions which passed filtering :
oligoSequences.txt contains the sequences - this file can be sent to the oligonucleotide synthesis facility CCseqBasicFragmentFile.txt is the ready-made parameter file for CCseqBasic analysis tool (to be used for sequenced fastq files of each sample) CaptureCompare_parameters.txt is the ready-made parameter file for CaptureCompare analysis tool (to compare 3+3 replicates of each sample)
Here the above files for the 30g test set - after CapSequm run, filtered with default parameters :
Or load the .zip containing all the above :