CCseqBasic
~ Output folders and files
(i) PREPARE the data for analysis
(ii) RUN THE ANALYSIS SEVERAL TIMES ,
generate useful visualisations and reports on the fly
(iii) COMBINE THE FINAL RESULTS ,
and generate a summary figure
Details below
The (i-iii) order is also the order of the output folders of the pipeline :
(i) PREPARE the data for analysis
- Folder F1_beforeCCanalyser
(ii) RUN THE ANALYSIS SEVERAL TIMES ,
generate useful visualisations and reports on the fly
- Folders F2-F5 like so :
F2_redGraphs
run the analysis once, generate the "red graphs" of the UCSC data hub
F3_orangeGraphs
run the analysis once, generate the "orange graphs" of the UCSC data hub
F4_blatPloidyFiltering ( and its results folder F5_greenGraphs_separate )
run the analysis once, generate the "green graphs" of the UCSC data hub
(iii) COMBINE THE FINAL RESULTS ,
and generate a summary figure
- Folders F6-F7 like so :
F6_greenGraphs_combined
combine the "flashed" and "nonflashed" green graphs,
run the analysis once, generate the "combined green graphs" of the UCSC data hub
F7_summaryFigure
generate a summary figure,
to make it easier to troubleshoot the above workflow
(i) PREPARE the data for analysis
- Folder F1_beforeCCanalyser
F1_beforeCCanalyser
preparing the sample for the analyserMappedReads.pl script this includes : read trimming, read combining (when R1 and R2 overlap), cutting reads on found restriction enzyme cut sites, read mapping
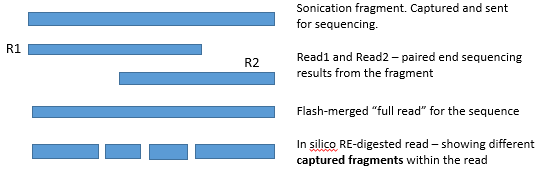
READ COMBINING :
"Flashed read" : program called "flash" is used to
combine the reads (when R1 and R2 overlap) to a single read
"Flashed reads" are the reads the program was able to combine to
a single entity (overlap of R1 and R2 reads was found)
R1
|------------
----------------| R2
|-------------------| flashed read (combining R1 and R2 to a single entity)
"Non-flashed read" : program called "flash" is used to
combine the reads (when R1 and R2 overlap) to a single read
"Non-flashed reads" are the reads the program was not able to combine to a single entity
(no overlap of R1 and R2 reads was found)
This means : there was no overlap, or the overlap was not "convincing enough"
(contained a lot of mismatches, or was very short overlap)
R1
|---------
-----------| R2
|--------- -----------| non-flashed read (R1 and R2 cannot be combined,
and continue to analysis as separate entities)
These two read sets : "flashed" and "non-flashed" are analysed separately
(all result files appear "twice" - once for the flashed read set, and once for the non-flashed one)
Thus, the output files have one Resriction enzyme cut output file for
"flashed" reads, and one for "non-flashed" reads.
The same goes for bowtie (mapping) results in the .bam files, as well as all subsequent steps.
(ii) RUN THE ANALYSIS SEVERAL TIMES , generate useful visualisations and reports on the fly
Each analysis round takes in the RE-digested, mapped, reads, and finds the captured sites, and the sites they interact with. Round after round more reads are filtered out of results (as they are duplicates or artifacts)
Folders F2-F5 like so :
F2_redGraphs
Run the analysis once, generate the "red graphs" of the UCSC data hub
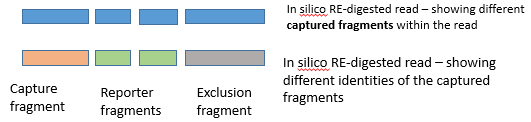
The fragments are assigned to be "captures" or "reporters" or "exclusion fragments".
Nothing is filtered.
F3_orangeGraphs
Run the analysis once, generate the "orange graphs" of the UCSC data hub
Duplicates are filtered
F4_blatPloidyFiltering
Filtering out problematic genomic regions (partial homology-based false positive signal,
repeat-region and genome build related issues).
Feeding this filtered data to further analysis run in folder F5_greenGraphs_separate, wherein
Run the analysis once, generate the "green graphs" of the UCSC data hub
These are now fully filtered files
This generates two sets of output files : 1) flashed and 2) non-flashed reads analysis results
( see above, chapter "READ COMBINING" )
Details below
Here more details and illustrations
about assigning fragments to "captures" and "reporters", like this :
 ( i.e. marking the mapped fragments to reflect the true results of the NG-CaptureC experiment )
( i.e. marking the mapped fragments to reflect the true results of the NG-CaptureC experiment )
Before reading the text below , check the link above : browse through the illustrations -
to get a clear picture what the text below refers to !
F2_redGraphs : run the analysis once, generate the "red graphs" of the UCSC data hub
When generating folder F2_redGraphs, the code reads in the mapped .bam files from F1_beforeCCanalyser folder
(one file for "flashed" and one for "non-flashed" reads).
The code will then run the "CCanalyser" code analyseMappedReads.pl for the first time.
This program takes the mapped fragments, and :
0) Re-unites the fragments to form the sonicated reads (which fragments belong together)
Read" and "fragment" : see the "illustrations" link above !
1) Checks if there is capture site(s) amongst the fragments of that read.
"Read" and "fragment" : see the "illustrations" link above !
If capture site is found, continue with the analysis of that read, otherwise skip that read.
2) Checks if there is at least one reporter fragment amongst the fragments of that read.
"Reporter fragment" : see the "illustrations" link above !
If reporter fragment(s) is found, continue with the analysis of that read, otherwise skip that read.
3) Checks if there is ONLY ONE capture site (not from many different captures)
among the fragments of that read.
If many different capture sites are found, skip that read, otherwise continue with the analysis of that read.
4) Checks if the read contains exactly the same mapped fragments, than
a previously analysed read (is a true read duplicate).
Not filtering duplicates in F2 folder yet - only updating the counters of them.
All the above is done separately for the flashed and non-flashed reads
(see above, chapter "READ COMBINING")
Thus, the F2 output folder contains non-filtered results for the
(i) flashed reads and (ii) non-flashed reads,
i.e each capture site gets 2 sets of files :
FLASHED_* and NONFLASHED_* output file sets.
Both of these sets contain, for each capture site :
(1) bam file (printout of all fragments(*) for that specific capture site
- from reads containing at least 1 capture and 1 reporter, in BAM format).
(2) windowed-wig file (the same data as the bam file, but in UCSC genome browser supported wig format,
windowed over 300 bases with 30 base sliding window, to show the signal better.)
(3) gff file (counts of reporter fragments(*), in restriction enzyme cut site bins, for whole genome.
For each read, each RE fragment is counted only once (if multiple fragments in the read
report same RE fragment, it is still counted only once)
Listing only RE fragment bins where non-zero amount of reporter fragments).
(4) wig file (the same data as the gff file, but in UCSC genome browser supported wig format.)
( (*)"Read" and "fragment" : see the "illustrations" above ! )
In addition to the above, both "flashed" and "nonflashed" get summary files :
(1) bam file (printout of all fragments for all capture sites
- from reads containing at least 1 capture and 1 reporter, in BAM format,
This file lists in the CO: column of sam file, which exact capture the read contained,
and whether the fragment in question was Reporter/Capture/Exclusion fragment,
whether the reporter was Cis or Trans, whether the fragment originates from Duplicated read).
(2) report file ( helpful summary of the counts of the reads and fragments along the analysis.
to interpret this, see this document :
http://userweb.molbiol.ox.ac.uk/public/telenius/captureManual/reportInterpret.txt
The output of the F2 folder is visualised in the UCSC data hub,
as separate "flashed" and "nonflashed" tracks for each capture oligo.
The F2 folder data is the red graph of the UCSC data hub (thus, the output folder is called F2_redGraphs)
F3_orangeGraphs : run the analysis once, generate the "orange graphs" of the UCSC data hub
The output sam file (where all the reads where at least one capture and one reporter) of F2_redgraphs,
enters another round of analyseMappedReads.pl
The analysis and output files is exactly the same as in F2_redgraphs, except step (4) :
The program takes the F2 folder output sam reads, and :
0) Re-unites the fragments to form the sonicated reads (which fragments(*) belong together)
1) Checks if there is capture site(s) amongst the fragments(*) of that read.
2) Checks if there is at least one reporter fragment amongst the fragments(*) of that read.
3) Checks if there is ONLY ONE capture site among the fragments(*) of that read.
4) Checks if the read contains exactly the same mapped fragments(*), than
a previously analysed read (is a true read duplicate).
Filtering duplicates - printing only reads(*) which are not PCR duplicates.
Again this step is done separately for the flashed and nonflashed reads,
The output files are exactly the same as in F2_redgraphs :
For each capture site, for both FLASHED and NONFLASHED reads :
(1) bam file (printout of all fragments(*) for that specific capture site
(2) windowed-wig file (the same data as the bam file 300 bases window with 30 bases slide)
(3) gff file (counts of reporter fragments(*) in restriction enzyme cut site bins)
(4) wig file (the same data as the gff file, but in wig format.)
Summary files (for "flashed" and "nonflashed" reads) :
(1) bam file (printout of all fragments for all capture sites, with CO: column of fragment(*) identity details)
(2) report file ( summary of the counts of the reads and fragments along the analysis )
( (*)"Read" and "fragment" : see the "illustrations" above ! )
The output of the F3 folder is visualised in the UCSC data hub,
as separate "flashed" and "nonflashed" tracks for each capture oligo.
The F3 folder data is the orange graph of the UCSC data hub (thus, the output folder is called F3_orangeGraphs)
F4_blatPloidyFiltering ( and its results folder F5_greenGraphs_separate ) : filter the files for homology-based artifacts and genome build based artifacts, run the analysis again
The output sam file (where all the reads where at least one capture and one reporter) of F3_orangegraphs,
enters filtering steps.
Again this step is done separately for the flashed and nonflashed reads.
BLAT-filtering (removing false positive hits due to homologous regions ) :
Whole genome homology-finder tool BLAT is used to screen for false positive long-range interaction regions.
This is done to avoid embarrasment : sometimes long range cis and trans interactions can be seen
just because the capture site happens to resemble closely ANOTHER location in the genome
(and mismapped reads cause this to show up as a long range "true" interaction).
This filtering is done like so :
Each capture site ( i.e. the whole RE fragment where the capture site resides + its "exclusion zone"(*))
is suspected to a BLAT run, where this site is mapped against the whole genome.
If hits are found (i.e. if the site has homologous sites elsewhere in the genome),
+/- 20 000 bases from each hit are eliminated as reporter fragments
(except hits +/- 200 000 bases from the target site itself, to not to filter short-range cis contacts).
( (*)"Exclusion zone" : see the "illustrations" above ! )
Thus, the sam files of each oligo in turn are searched for these "to-be-eliminated" regions,
and if reads mapping there are found, the reporter fragments in there are deleted.
Used blat parameters are :
minMatch=2 tileSize=11 maxIntron=4000 stepSize=5 minScore=10
(and can be modified with user-given flags)
The default parameters mean that the program searches for
11-base wide homology regions (full matching 11 bases minimum)
and if it finds at least 2 of these
separated separated by maximum 4000 bases from each others,
this triggers a homologous region - and all of these are to be removed in the blat-filtering step.
Step size determines how often this "search for homology" is restarted. Here we do in 5 bases steps
along whole genome.
Min score triggers "how similar" the sequences need to be to trigger a homologous region.
Value 10 (used here) flags regions as homologous relatively easily.
Generating the lists of the to-be-filtered regions is time consuming,
and generating these lists by running CCseqBasic with --onlyBlat flag
before running the actual analysis runs, is recommended.
The --onlyBlat run homology lists can be re-used for all designs using the same oligos,
such as replicates of a sample, or different cell types captured with same oligos).
Individual oligos can be re-used as well (to avoid re-running homology searches
for oligos shared between designs).
Blacklist-filtering ( removing genome-build based artifacts ) :
The sam files are also subjected to filtering of "blacklisted regions".
These are generally very repeat-rich regions, which generate deep (but artifactual) signals in the data.
These "false positive" peaks are seen in all samples, in all data types (ChIP-seq, ATAC-seq, RNA-seq, captureC),
and if some of these regions are within the interaction region of any of the capture probes,
convincing-looking interaction signal can result, and this may lead to false interpretation of the data.
If you are interested in repeat-rich regions, you may wish to turn this filter off,
by running CCseqBasic with --noBlacklistFilter
The F4_blatPloidyFiltering folder contains the various log files and the
to-be-filtered-regions listings of the above two filters.
It also contains the sam files (originally from F3_orangeGraphs),
after they have been filtered for the found artifact regions.
F5_greenGraphs_separate : run the analysis once, generate the "green graphs" of the UCSC data hub
The filtered output sam files of F4_blatPloidyFiltering,
enter another round of analyseMappedReads.pl
The analysis and output files is exactly the same as in F2_redgraphs and F3_orangegraphs.
( this analysis is not exactly needed any more - but provides
useful read counter data and visualisations in the end of the run)
Again this step is done separately for the flashed and nonflashed reads,
The output files are exactly the same as in F2_redGraphs and F3_orangeGraphs :
For each capture site, for both FLASHED and NONFLASHED reads :
(1) bam file (printout of all fragments(*) for that specific capture site
(2) windowed-wig file (the same data as the bam file 300 bases window with 30 bases slide)
(3) gff file (counts of reporter fragments(*) in restriction enzyme cut site bins)
(4) wig file (the same data as the gff file, but in wig format.)
Summary files (for "flashed" and "nonflashed" reads) :
(1) bam file (printout of all fragments for all capture sites, with CO: column of fragment(*) identity details)
(2) report file ( summary of the counts of the reads and fragments along the analysis )
( (*)"Read" and "fragment" : see the "illustrations" above ! )
The output of the F5 folder is visualised in the UCSC data hub.
The F5 folder data is the green graph of the UCSC data hub (thus, the output folder is called F5_greenGraphs)
This is the last folder, where the data is presented separately for "flashed" and "nonflashed" reads.
This is why the folder name is F5_greenGraphs_separate
(iii) COMBINE THE FINAL RESULTS ,
and generate a summary figure
Folders F6-F7 like so :
F6_greenGraphs_combined
combine the "flashed" and "nonflashed" green graphs,
run the analysis once, generate the "combined green graphs" of the UCSC data hub
F7_summaryFigure
generate a summary figure,
to make it easier to troubleshoot the above workflow
This generates the final set of output files : COMBINED "flashed" + "non-flashed" reads analysis results
Details below
F6_greenGraphs_combined : combine the "flashed" and "nonflashed" data, run the analysis once, generate the final combined "green graph" of the UCSC data hub
The sam files of "flashed" and "nonflashed" reads from F5_greenGraphs_separate are combined,
and this combined data enters the (last and final) round of analyseMappedReads.pl
The analysis and output files is exactly the same as in
F2_redGraphs and F3_orangeGraphs and F5_greenGraphs_separate.
( this analysis is not exactly needed any more - but provides
useful read counter data and visualisations in the end of the run)
The output files are exactly the same as before - but now the FLASHED and NONFLASHED data are not separate,
but combined in the COMBINED files :
(1) bam file (printout of all fragments(*) for that specific capture site
(2) windowed-wig file (the same data as the bam file 300 bases window with 30 bases slide)
(3) gff file (counts of reporter fragments(*) in restriction enzyme cut site bins)
(4) wig file (the same data as the gff file, but in wig format.)
Summary files (for "flashed" and "nonflashed" reads) :
(1) bam file (printout of all fragments for all capture sites, with CO: column of fragment(*) identity details)
(2) report file ( summary of the counts of the reads and fragments along the analysis )
( (*)"Read" and "fragment" : see the "illustrations" above ! )
The output of the F6 folder is visualised in the UCSC data hub, one track for each capture oligo.
The F6 folder data is the green COMBINED graph of the UCSC data hub
(thus, the output folder is called F6_greenGraphs)
F7_summaryFigure : easy-to-interpret output counters visualisation, to go with the UCSC data hub
The above data flow can be quite exhausting to troubleshoot, so the reporter counter files are transformed to easy-to-interpret graphical representation.
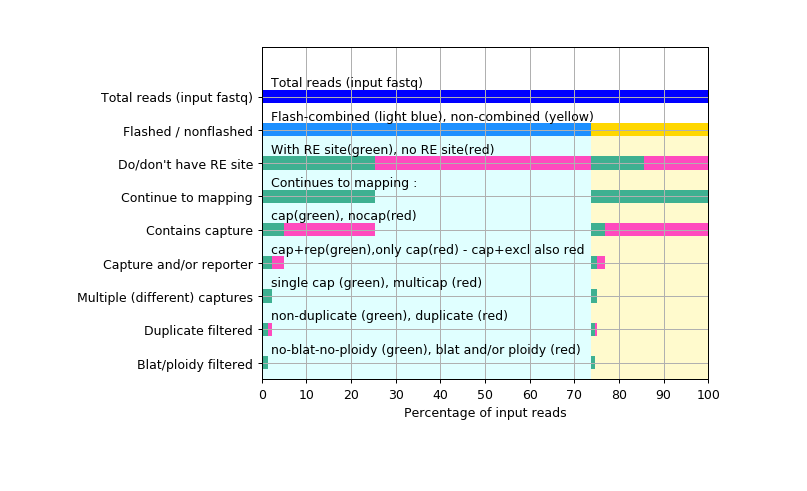
This visualisation file is available in the UCSC data hub by clicking any of the data tracks,
or opening "track preferences" page by right-clicking the track
This folder contains the summary figure, and the log files about making it.