CCseqBasic
~ Generate capturesite (RE fragment) coordinate file
Before you can start your pipeline run, you have to generate a capturesite (RE fragment) coordinate file.
Your CapSequm output should have this file already - it was saved under name CCseqBasicFragmentFile
The capturesite (RE fragment) coordinate file lists the DpnII (or other Restriction Enzyme) fragments, within which your capture oligos reside.
( these are 1-based coordinates - like f.ex. gff and sam files, not 0-based like f.ex. bed files.
More about 1-based and 0-based files f.ex. in
here
)
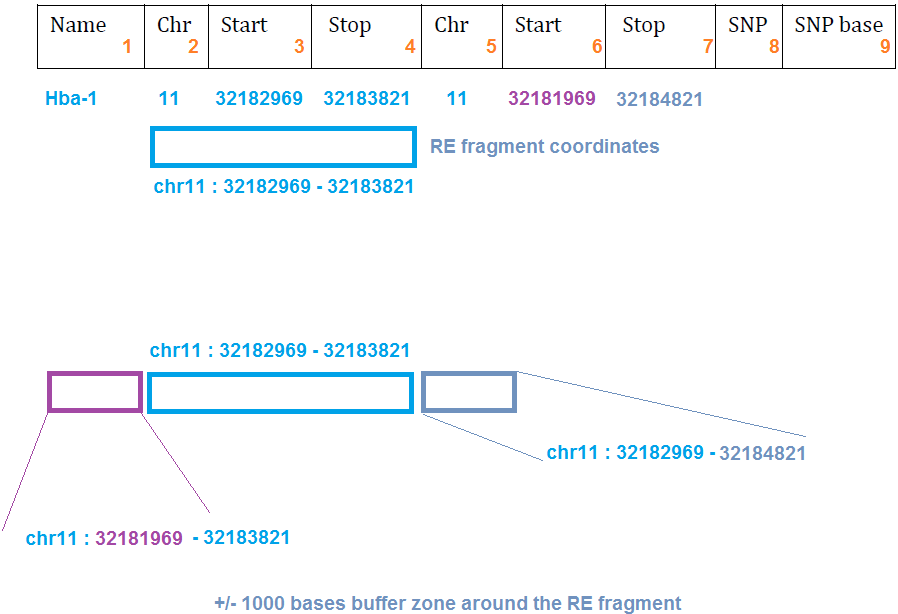
The "buffer zone" is called Exclusion Zone, and it is needed for determining how wide area around the capture RE fragment should be removed due to incomplete RE digestion in the experiment - to avoid reporting incomplete digestion as true proximity contact signal.
The actual width of the exclusion zone can be finetuned, but a good starting point is +/- 1000 bases both directions.
If you are generating capturesite (RE fragment) file by hand, instead of using the readymade file from CapSequm, please read these instructions carefully, to stay faithfully in correct coordinate set, to allow CCseqBasic to recognise the captured fragments properly. If the given coordinates for the capture RE fragments are even one base too NARROW, significant losses of reported interactions will result. Giving them too wide, on the other hand, will not matter at all.
When generating capturesite file by hand, remember to list each of the captured DpnII fragments only once (even when you captured from two oligos within the fragment). If the RE fragment is listed twice - it is seen as "double capture" (each read mapping to more than one capture site). This kind of reads are eliminated, and all reads originating from that capture location are filtered out in CCseqBasic run, essentially making the whole capture site "vanish" from the results.
If you are interested in using the special "SNP run style" related columns (to be combined with run flag --snp ), you need to fill the last 2 columns of the file too :
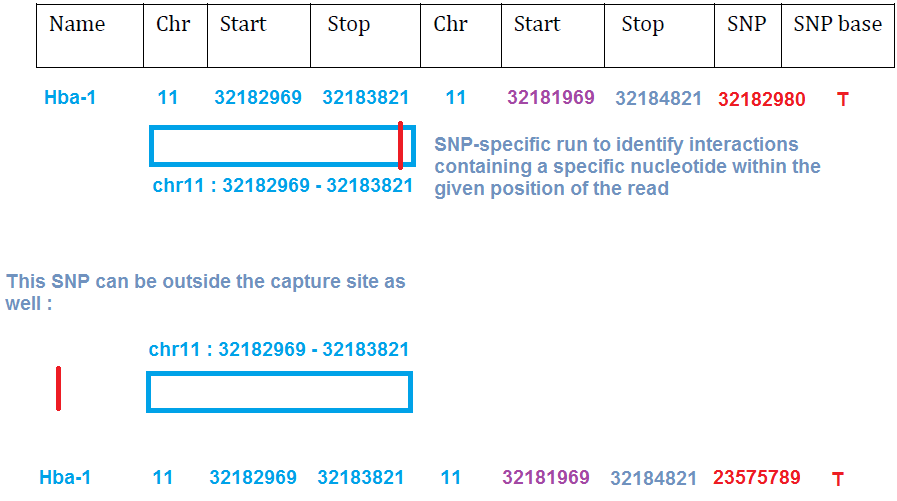
More information of the exact format of the CCseqBasicFragmentFile , and instructions to generate one manually : Generating capturesite (RE fragment) file by hand